Research
Research Interests
Our research focuses on the biosynthetic mechanisms and metabolic engineering of novel antifungal and anti-MRSA antibiotics from underexplored bacteria, as well as mycotoxins and other bioactive natural products from plant pathogenic fungi and endophytes.
Current Research
- Investigating the molecular mechanisms underlying the biosynthesis of novel antibiotics, mycotoxins, and other bioactive natural products.
- Discovering new antibiotics and signaling molecules from Lysobacter bacteria, endophytic fungi, and other underexplored sources.
- Engineering metabolic pathways to enhance the production of valuable natural products.
Research in the Du group is at the interface of chemistry and biology, focusing on understanding how microorganisms biosynthesize structurally complex and biologically active natural products. Our ultimate goal is to harness this knowledge for the development of new bioactive compounds through biosynthetic approaches. Our research integrates expertise from chemistry, biochemistry, chemical biology, and synthetic biology. Currently, we are pursuing two major projects aimed at advancing antibiotic discovery and metabolic pathway engineering.
New antibiotics from underexplored microorganisms
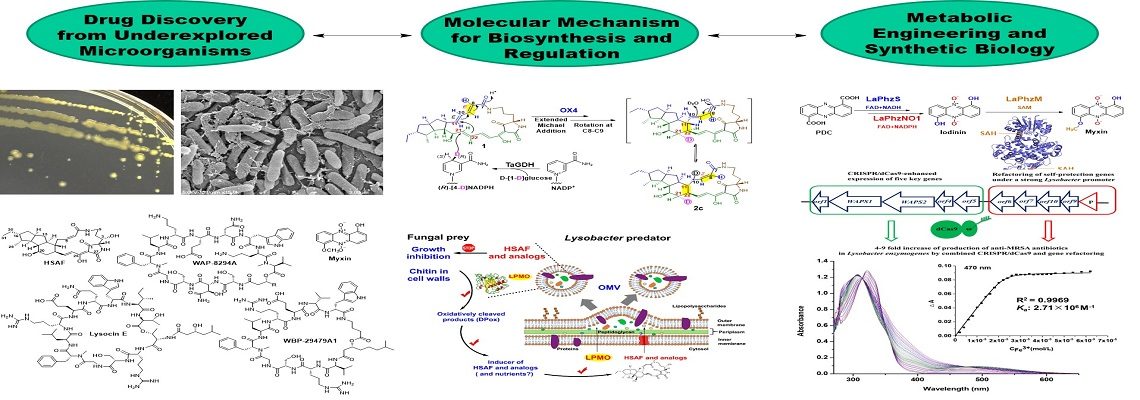
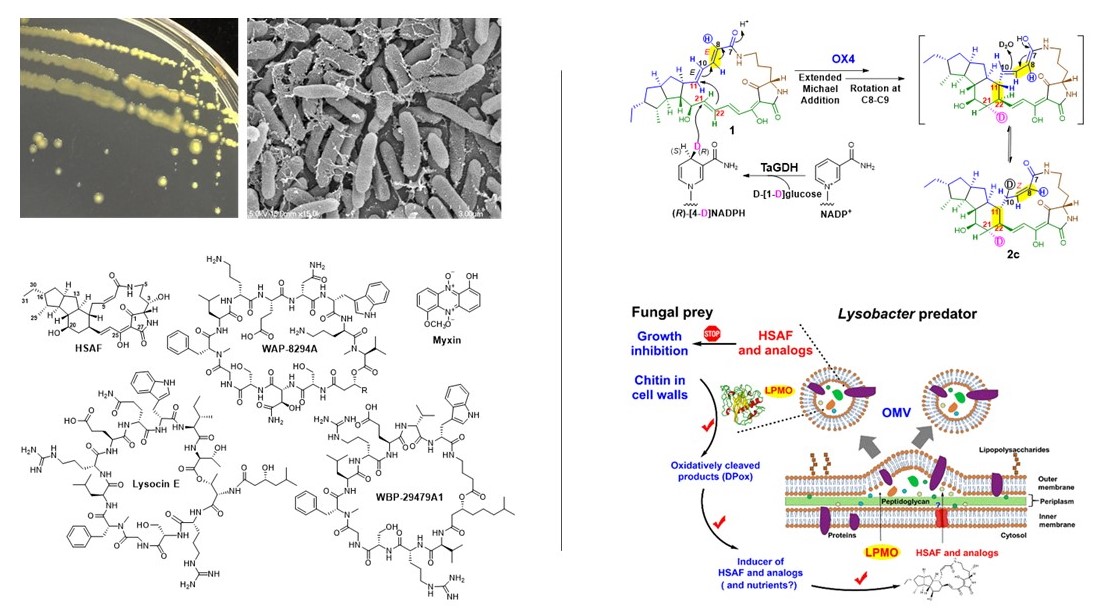
Lysobacter is a genus of environmental bacteria first classified in 1978, with several species recognized as prolific producers of bioactive natural products and emerging biocontrol agents (Nat. Prod. Rep. 2012; Nat. Prod. Rep. 2022). From the biocontrol agent Lysobacter enzymogenes, we have isolated HSAF, a potent antifungal compound with a novel mode of action (Mol. Biol. Cell 2006; RSC Adv. 2016; Biochim. Biophys. Acta 2016) and unique structural features distinct from existing fungicides and antifungal drugs (Antimicrob. Agents Chemother. 2007). We identified the genes responsible for HSAF biosynthesis and uncovered a previously unrecognized hybrid polyketide-peptide biosynthetic mechanism (J. Am. Chem. Soc. 2011; Biochemistry 2012; Angew. Chem. Int. Ed. 2014, 2018; Biochemistry 2020).
We have isolated three classes of potent anti-MRSA cyclic lipodepsipeptides and identified their biosynthetic gene clusters (Antimicrob. Agents Chemother. 2011; ACS Synth. Biol. 2013, 2018, 2020; Org. Lett. 2019). These cyclic lipopeptides are structurally distinct from daptomycin, a clinically used antibiotic effective against Gram-positive pathogens, including MRSA and vancomycin-resistant enterococci (VRE).
Through genome mining, we discovered various bioactive natural products, including the broad-spectrum antibiotic phenazines, and identified an unprecedented Baeyer-Villiger-like flavin enzyme responsible for phenazine N-oxidation (Org. Lett. 2016, 2017; ACS Chem. Biol. 2018).
Despite its promise, Lysobacter has remained challenging to manipulate genetically. To address this, we have developed novel tools for activating silent biosynthetic genes and advancing synthetic biology in Lysobacter, enabling improved compound yields and unveiling intricate regulatory mechanisms (Appl. Environ. Microbiol. 2017, 2021; ACS Chem. Biol. 2021; J. Agric. Food Chem. 2023, 2025).
Mycotoxins from Food-Borne Molds
Our research aims to develop innovative strategies for reducing or eliminating mycotoxins in food and feed. We have focused on fumonisins, a group of fungal polyketides, to understand their biosynthetic mechanisms. This knowledge will facilitate rational approaches to controlling mycotoxin contamination.
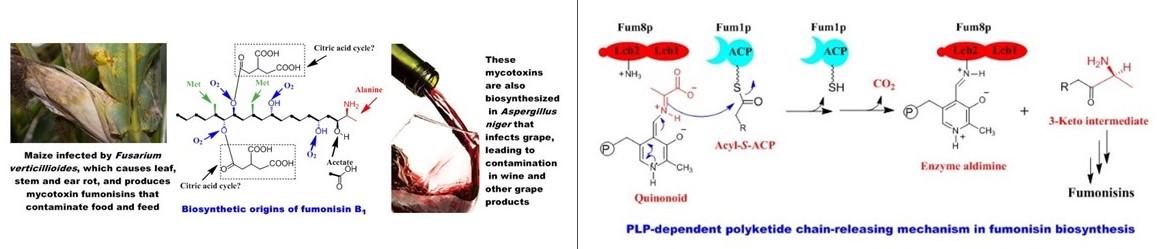
Fumonisins rank among the five most economically significant mycotoxins in agriculture and the food industry (Biopolymers 2010). They are produced by the pathogenic fungus Fusarium verticillioides, a widespread contaminant of corn and maize-derived food and feed. Consumption of fumonisin-contaminated corn leads to fatal diseases in livestock and poses a cancer risk to humans.
To investigate fumonisin biosynthesis, we developed a genetic system capable of precisely modifying biosynthetic genes in filamentous fungi. Using this system, we created gene-specific mutants, identified biosynthetic intermediates, and established a biosynthetic pathway (J. Am. Chem. Soc. 2007). Additionally, we leveraged E. coli and Saccharomyces cerevisiae as heterologous hosts to express and study fungal genes (Biochemistry 2006).
Our studies uncovered an unprecedented PLP-dependent polyketide chain-releasing mechanism, in which a discrete 2-oxoamine synthase catalyzes a decarboxylative condensation between L-alanine and acyl-S-ACP (J. Am. Chem. Soc. 2009). This reaction facilitates polyketide chain termination and offloading while simultaneously introducing a new carbon-carbon bond and an amino group. Unlike the thioesterase/cyclase-mediated chain release observed in bacterial and other fungal polyketide biosynthesis, this mechanism represents a novel paradigm in fungal secondary metabolism (Nat. Prod. Rep. 2010).