Research Area 1: Biosensor Design
The focus of this research area is the design and fabrication of folding- and dynamics-based electrochemical biosensors. The biorecogntion elements commonly used in the fabrication of these sensors include DNA, RNA, aptamers, and peptides. This sensing platform is highly versatile, a wide range of target analytes, including DNA, small molecules, proteins and antibodies, have been detected using this sensing strategy. Unlike most biosensors, these sensors are reagentless, reusable, rapid, sensitive, specific, and selective enough to be employed directly in realistically complex media such as whole blood, urine, saliva, soil extract and various food matrices.
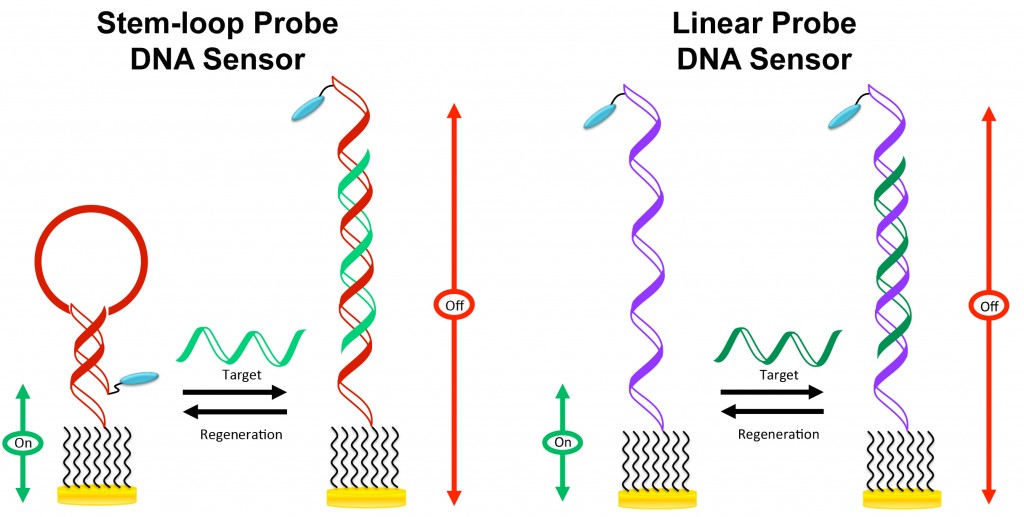 The two well-characterized “signal-off” DNA sensors are the stem-loop probe (SLP) and linear probe (LP) electrochemical DNA (E-DNA) sensors. Both sensors utilize a thiolated DNA probe that is modified with a redox label such as methylene blue (MB).
The two well-characterized “signal-off” DNA sensors are the stem-loop probe (SLP) and linear probe (LP) electrochemical DNA (E-DNA) sensors. Both sensors utilize a thiolated DNA probe that is modified with a redox label such as methylene blue (MB).
LEFT: For the SLP sensor, in the absence of the target DNA, the MB label is positioned close to the electrode surface because of the probe conformation. In the presence of the target DNA, flexibility of the MB-labeled probe is severely hindered, dampening the electron transfer kinetics. A large reduction in MB current is thus observed in both alternating current voltammetry and cyclic voltammetry.
RIGHT: For the LP sensor, the probe is highly flexible in the absence of the target, target binding induces a change in probe dynamics, giving rise to a large reduction in the MB current.
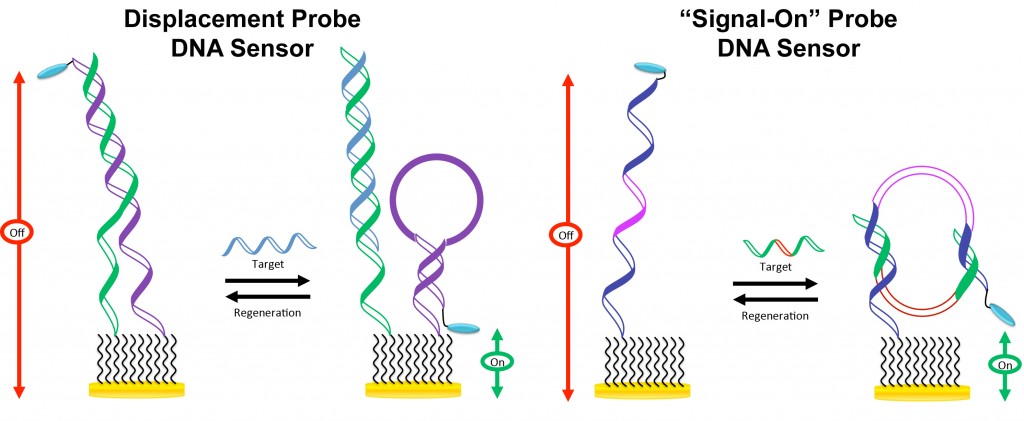
Two “signal-on” E-DNA sensors have been developed recently.
LEFT: The “displacement-based” E-DNA sensor utilizes two surface-immobilized DNA probes that are partially hybridized to each other in the absence of the target. In this case, the electron transfer kinetics is dampened because of probe rigidity. In the presence of the target DNA that binds to the unlabeled capture probe, the inter-probe duplex is disrupted, releasing the MB-labeled signaling probe that is capable of forming a hairpin structure. Formation of the hairpin structure positions the MB label close to the electrode, giving rise to an increase in the MB current.
RIGHT: This “signal-on” E-DNA sensor is fabricated using a linear DNA probe containing a flexible oligothymine linker. The probe is flexible in the absence of the target, target binding, however, induces formation of a “close” structure, positioning the MB label close to the electrode, resulting in an increase in the MB current.
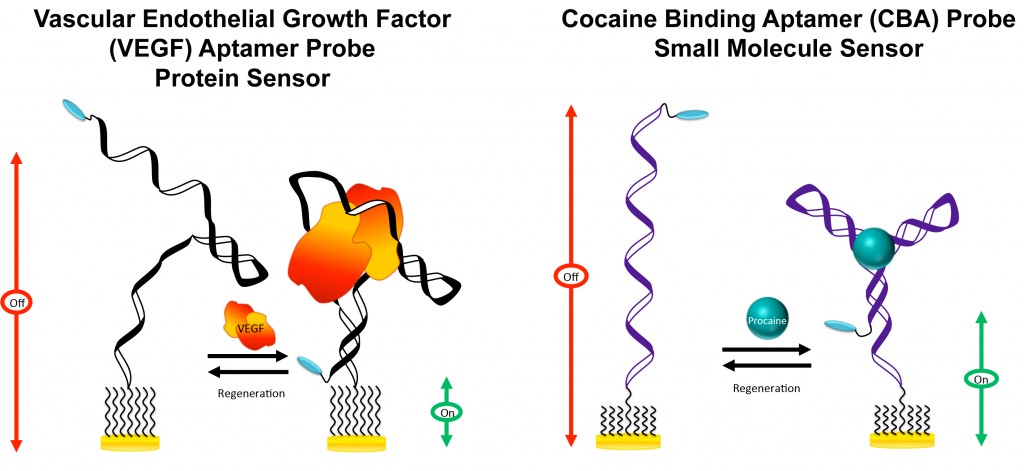
Two “signal-on” electrochemical aptamer-based (E-AB) sensors have been developed recently.
LEFT: In the absence of the target (VEGF), the aptamer probe is partially unfolded. Upon binding to the target, the probe forms a “close” structure, resulting in a change in the electron transfer kinetics and an increase in the MB current.
RIGHT: In the absence of the target (cocaine), the aptamer probe is fully or partially extended depending on the ionic strength of the solvent. Target binding induces formation of a unique “three-way junction”, leading to a change in the electron transfer kinetics, which is signified by an increase in the MB current.
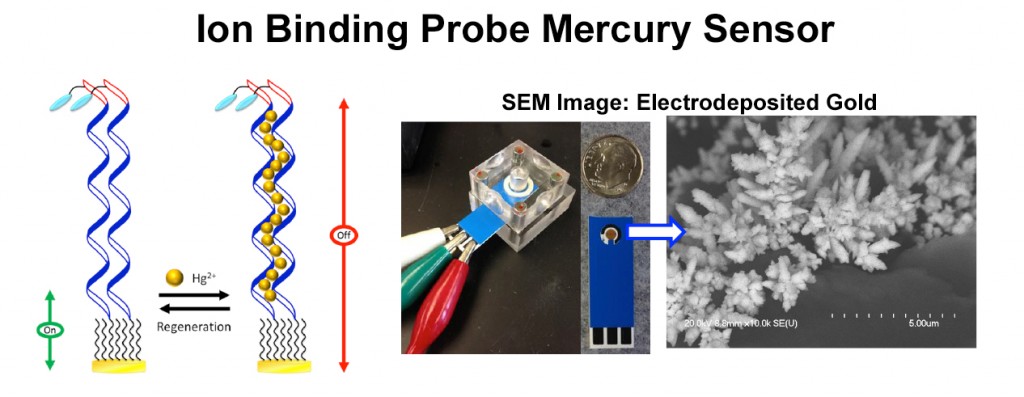 For the electrochemical ion (E-ION) Hg(II) sensor, in the absence of the target, the thymine (T)-containing DNA probes are highly flexible, enabling efficient electron transfer between the electrode and the MB label. Binding of Hg(II) via the well-characterized T-Hg(II)-T motif rigidifies the probes, resulting in a large reduction in the MB current. This sensor has been demonstrated to be functional even when fabricated on a gold-plated screen-printed carbon electrode. This single-use, paper-based sensor is ideal for Hg(II) detection in environmental and food samples.
For the electrochemical ion (E-ION) Hg(II) sensor, in the absence of the target, the thymine (T)-containing DNA probes are highly flexible, enabling efficient electron transfer between the electrode and the MB label. Binding of Hg(II) via the well-characterized T-Hg(II)-T motif rigidifies the probes, resulting in a large reduction in the MB current. This sensor has been demonstrated to be functional even when fabricated on a gold-plated screen-printed carbon electrode. This single-use, paper-based sensor is ideal for Hg(II) detection in environmental and food samples.
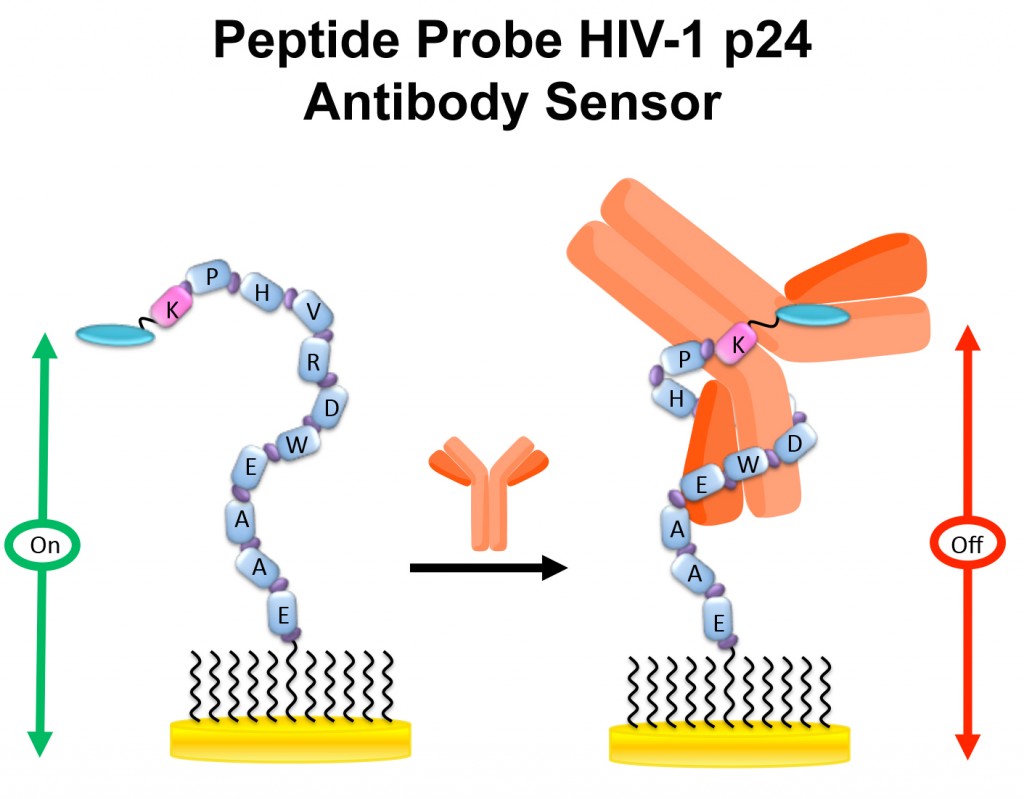
Several electrochemical peptide based (E-PB) sensors have been developed to date. Unlike the E-DNA, E-AB and E-ION sensors, fabrication of these sensors requires the use of MB-labeled peptide probes.
For the E-PB HIV sensor, the peptide probe is highly flexible in the absence of the target antibody (e.g., HIV anti-p24 antibody), binding of the target antibody changes both probe conformation and flexibility, resulting in a significant reduction in the MB current. While the signaling mechanism of the E-PB sensor is similar to the LP E-DNA sensor, the use of peptide recognition probes enables detection of a wider selection of disease biomarkers.
These sensors, owing to their unique properties which include high sensitivity, specificity and selectivity, can potentially be used as an alternative to immunoassays commonly used in disease diagnosis.
Recent Publications
– Use of Thiolated Oligonucleotides as Anti-fouling Diluents in Electrochemical Peptide-based Sensors. McQuistan, A.; Zaitouna, A. J.; Echeverria, E.; Lai, R. Y., Chem. Commun., 2014, 50, 4690-4692.
– Effect of Redox Label Tether Length and Flexibility on Sensor Performance of Displacement-based Electrochemical Nucleic Acid Sensors. Yu, Z.; Zaitouna, A. J.; Lai, R.Y., Analytica Chimica Acta, 2014, 812, 176-183.
– Characterization of an Electrochemical Mercury Sensor Using Alternating Current, Cyclic, Square Wave and Differential Pulse Voltammetry. Guerreiro, G. V.; Zaitouna, A. J.; Lai, R. Y. Analytica Chimica Acta, 2014, 810, 79-85.
– Electrochemical Techniques for Characterization of Stem-loop Probe and Linear Probe-based DNA Sensors. Lai, R. Y.; Walker, B.; Stormberg, K.; Zaitouna, A. J.; Yang, W. Methods, 2013, 64, 267-275.
– “Signal-on” Electrochemical DNA Sensor with an Oligo-thymine Spacer for Point Mutation Detection. Wu, Y.; Lai, R.Y. Chem. Commun. 2013, 49, 3422-3424.
– The Effect of Signaling Probe Conformation on Sensor Performance of a Displacement-based Electrochemical DNA Sensor. Yu, Z.; Lai, R.Y. Anal. Chem., 2013, 85, 3340-3346.
– Development of an Electrochemical Insulin Sensor Based on the Insulin-linked Polymorphic Region. Gerasimov, J.Y.; Schaefer, C.S.; Yang, W.; Grout, R.L.; Lai, R.Y. Biosensors and Bioelectronics, 2013, 42, 62-68.
