Affinity Mass Spectrometry
The combined use of affinity chromatography with mass spectrometry for bioanalysis is a topic that is of further interest to our group. These types of studies might involve the use of affinity chromatography as either a selective separation method prior to methods such as LC/MS, tandem mass spectrometry (e.g., LC/MS/MS), or matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. We also combine the use of affinity chromatography to provide information on the function of a biological molecule and its interactions with other compounds, while we use mass spectrometry to provide information on the structure of this biomolecule. This combination is a powerful set of tools for the examination of complex biological systems.
As an example, we were the first group to use mass spectrometry to examine the immobilization of proteins for use in affinity chromatography. During the immobilization of a protein it is generally desirable for this ligand to be attached to a support in a way that allows the ligand to closely retain its native structure and activity. Traditionally, these ligands are covalently immobilized to surfaces through amine-based coupling techniques such as the Schiff base method. To examine this process, we used a method based on quantitative proteomics, mass spectrometry, and 16O/18O-labeling to determine which of the many possible amine groups on HSA are involved in its immboilization and to determine the impact of immobilization through these sites. The general scheme for this labeling process is shown in the figure below. The immobilized HSA was digested with a proteolytic enzyme in the presence of normal water, while a soluble form of the same protein was digested in 18O-enriched water. After the digestion reactions were quenched, the soluble fractions of the digests were combined, fractionated, and analyzed using MALDI-TOF MS.
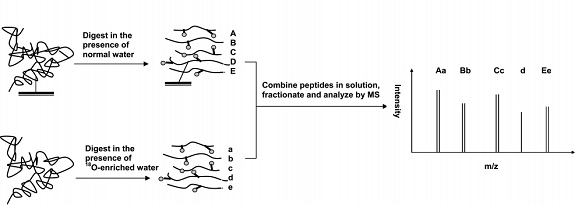
Use of quantitative proteomics and mass spectrometry to examine protein immobilization for affinity chromatography
This approach was able to identify the seven major immobilization sites for the protein human serum albumin (HSA) when using the Schiff base method, none of which were located in the major drug binding region of HSA. This result indicating that the immobilization of HSA under these conditions should not greatly alter the binding activity of this protein, as has also been observed experimentally in separate studies.
In another ongoing project in our group, we are using high-performance affinity chromatography (HPAC) and mass spectrometry to examine the changes in drug binding to serum proteins that occurs during diabetes. This work involves the use of both HPAC and MALDI-TOF MS or LC/MS/MS. This method has also been used to quantitatively compare HSA to glycated HSA, as is found in serum during diabetes. This work has provided insight into the specific regions on HSA where glycation is most apt to occur and has made it possible to assess the possible impact of these structural changes on the binding of drugs to HSA during diabetes. The results of this work should eventually lead to a better understanding on how drug activity may change between diabetic and non-diabetic patients and lead to better treatment regimes for diabetic patients based on personalized medicine.
Examples of Recent Work with Affinity Mass Spectrometry
- Chunling Wa, Ronald L. Cerny and David S. Hage, “Obtaining High Sequence Coverage in MALDI-TOF MS for Studies of Protein Modification: Analysis of Human Serum Albumin as a Model”, Anal. Biochem, 349 (2006) 229-241.
- Chunling Wa, Ronald L. Cerny and David. S. Hage, “Identification and quantitative studies of protein immobilization sites by stable isotope labeling and mass spectrometry” Anal. Chem., 78 (2006) 7967-7977.
- Chad J. Briscoe, William Clarke and David S. Hage, “Affinity Mass Spectrometry”, In: Handbook of Affinity Chromatography, (D. S. Hage, Editor), CRC Press/Taylor & Francis, 2006, Chapter 27.
- Chunling Wa, Ronald L. Cerny, William A. Clarke and David S. Hage, “Characterization of Glycated Adducts on Human Serum Albumin by MALDI-TOF MS”, Clin. Chim. Acta, 385 (2007) 48-60.
- Chad J. Briscoe and David S. Hage, “Factors Affecting the Stability of Drugs and Drug Metabolites in Biological Matrices”, Bioanalysis, 1 (2009) 205-220.
- Omar Barnaby, Chunling Wa, Ronald L. Cerny, William Clarke and David S. Hage, “Quantitative Analysis of Human Serum Albumin using 16O/18O-Labeling and Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry”, Clin. Chim. Acta, 411 (2010) 1102-1110.
