Immobilization and Characterization of Biochemicals
Part of our research involves the optimization or creation of new schemes for immobilizing and modifying biological agents. The immobilization of biological agents can be accomplished using three general approaches. These methods include noncovalent techniques like nonspecific and biospecific absorption, covalent coupling techniques, entrapment, and molecular imprinting. Students working in this area explore the use of both organic and inorganic methods of synthesis for the attachment and modification of biochemicals, as needed in high-performance affinity chromatography or other bioanalytical methods.
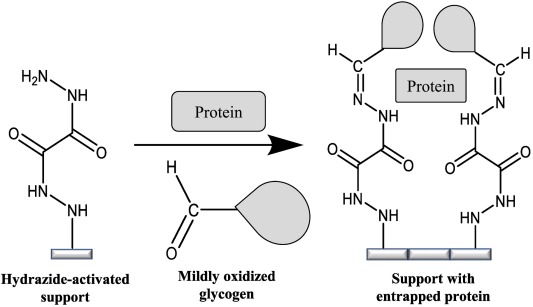
Fig: Entrapment of soluble protein using using a hydrazide-activated support and mildly oxidized glycogen.
We have used all of these methods in our group, but most of our work involves the use and development of covalent immobilization methods. Covalent immobilization can be initiated through several different functional groups on the ligand. Certain immobilization techniques, such as the cyanogen bromide method, the Schiff base method (i.e., reductive amination), the N-hydroxysuccinimide technique, and the carbonyldiimidazole method, use free amine groups during protein immobilization. Other functional groups that may be employed include sulfhydryl groups, carboxyl groups, and carbonyl groups. For instance, our group has developed a general method that can be used for the site-selective immobilization of antibodies through their carbohydrate chains. We have also optimized and studied techniques for the immobilization of the proteins human serum albumin (HSA), alpha 1-acid glycoprotein (AGP), and proteolytic enzymes. Several patents have been awarded to our group for this research and many papers have resulted for this work, as have appeared in journals like Analytical Chemistry, Analytical Biochemistry, Bioconjugate Chemistry and the Journal of Chromatography (A and B).
Another interesting type of immobilization that has recently been created and utilized in our group is an entrapment method. This entrapment method is a noncovalent technique for protein and ligand immobilization that is capable of being used with common HPLC supports. This entrapment method involves the physical containment of a protein or ligand in a polysaccharide-capped dihydrazide support. When these oxidized polysaccharides are incubated with the hydrazide-activated support, aldehyde groups on the oxidized polysaccharide will form a stable covalent bond with the hydrazide groups, a reaction known to occur even in a neutral aqueous buffer. This process will entrap the ligand in the support.
Our group also uses a variety of tools to study immobilized proteins as part of this work in the creation and optimization of immobilization methods. These tools have included protein assays, flow-injection analysis, infrared spectroscopy, solid-state nuclear magnetic resonance spectroscopy, and mass spectrometry. Both HPAC and capillary electrophoresis are also used as part of this work.
Examples of Recent Work in the Immobilization & Characterization of Biochemicals
1.) John Vargas-Badilla, Saumen Poddar, Shiden Azaria, Chenhua Zhang and David S. Hage*, “Optimization of Protein Entrapment in Affinity Microcolumns using Hydrazide-Activated Silica and Glycogen as a Capping Agent”, J. Chromatogr. B, 1121 (2019) 1-8. PMC6545136
2.) Chenhua Zhang and David S. Hage*, “Development and Evaluation of High Performance Lectin Microcolumns for Glycoform Analysis of Alpha1-Acid Glycoprotein”, Anal. Chim. Acta, 1078 (2019) 189-199.
3.) Zhao Li, Elliott Rodriguez, Shiden Azaria, Allegra Pekarek, and David S. Hage*, “Affinity Monolith Chromatography: A Review of General Principles and Applications”, Electrophoresis, 38 (2017) 2837-2850. (invited paper; top downloaded article for Electrophoresis (top 20) – 2017-2018).
4.) Xiwei Zheng, Zhao Li, Sandya Beeram, Ryan Matsuda, Erika L. Pfaunmiller, Maria Podariu, Christopher J. White II, NaTasha Carter, and David S. Hage*, “Analysis of Biomolecular Interactions using Affinity Microcolumns: A Review”, J. Chromatogr. B, 968 (2014) 49-63.