Prof. Guo
Dr. Guo received his B.S. and M.S. degrees in Organic Chemistry from Nankai University with Professor Jin-Pei Cheng, and his Ph.D. degree from Michigan State University with Professor John W. Frost. He pursued postdoctoral studies with Professor Peter G. Schultz at The Scripps Research Institute before joining the University of Nebraska – Lincoln, Department of Chemistry as an Assistant Professor in 2010.
University of Nebraska

Department of Chemistry
Since being founded in 1882, UNL’s Chemistry Department has lead in diverse areas from computational chemistry to synthetic organic chemistry, mass spectrometry and surface science — from chromatography and separations science to chemical biology and materials design, fabrication, and study.
Interfacing Chemsitry and Biology
My lab has two major research themes, Chemical Biology and Synthetic Biology. We use a multidisciplinary approach to tackle fundamental problems associated with human health and sustainable chemical production. We seek to translate fundamental scientific findings into biomedical and biotechnological applications.
A graduate student or postdoctoral researcher in my lab can expect to receive training in bioorganic chemistry, chemical biology, and synthetic biology, and significant exposure to one or more of the following specialties: organic synthesis, enzymatic synthesis, combinatorial (bio)chemistry, bio-molecular engineering, directed biomolecular evolution, unnatural amino acid mutagenesis, protein biochemistry, and molecular biology.
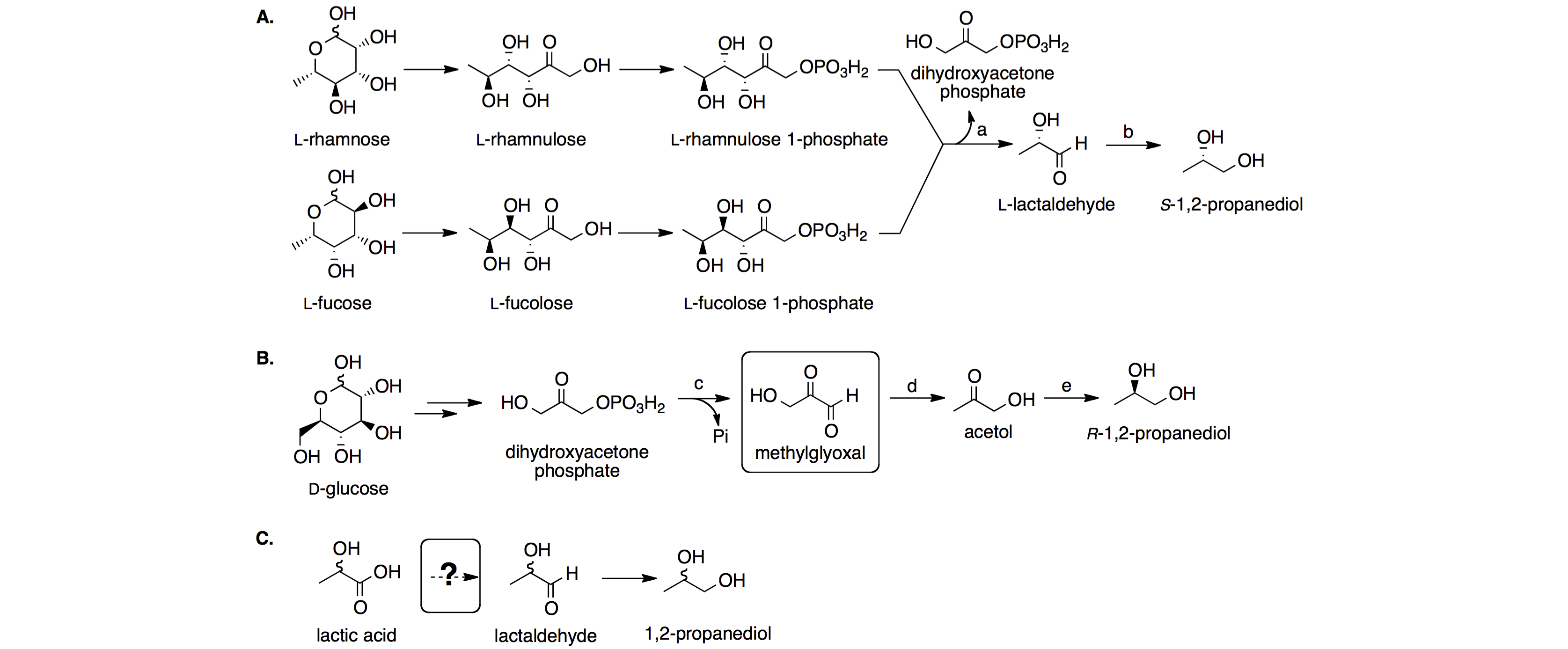
I. Genetic code engineering and unnatural amino acids
The genetic code specifies 20 common amino acids plus the rare occurrence of selenocysteine and pyrrolysine. Genetic incorporation of unnatural amino acids with novel physical, chemical, and biological properties can significantly enhance scientists’ ability to probe and manipulate protein structure and function. The goal of our research is to reprogram the universal genetic code, including both nonsense codon reassignment and quadruplet codon decoding, so that unnatural amino acid building blocks or exogenous regulatory elements can be readily introduced into biological systems.
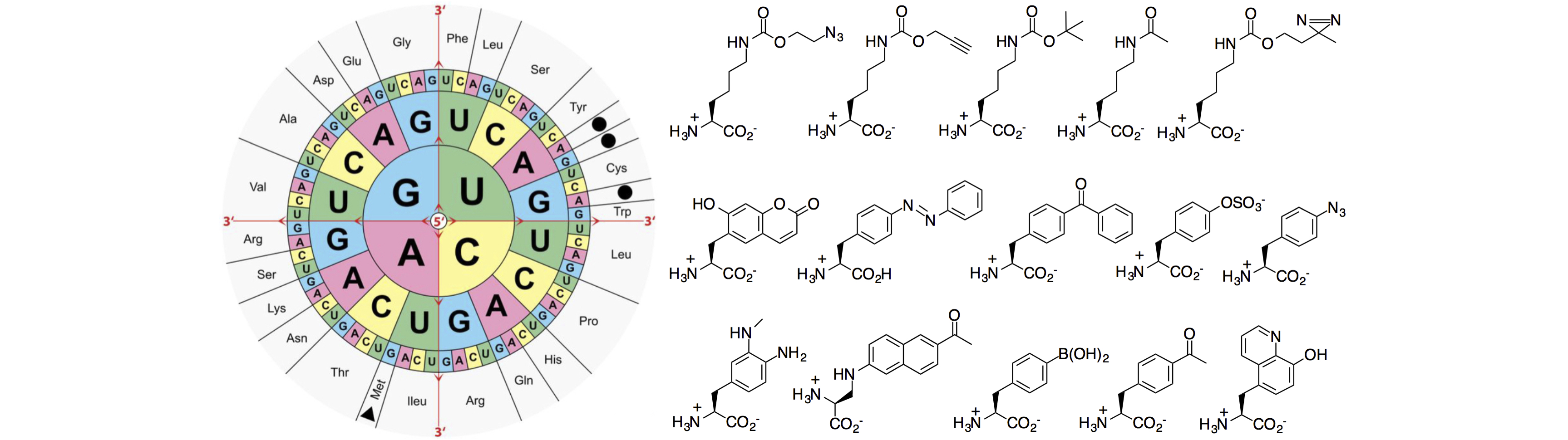
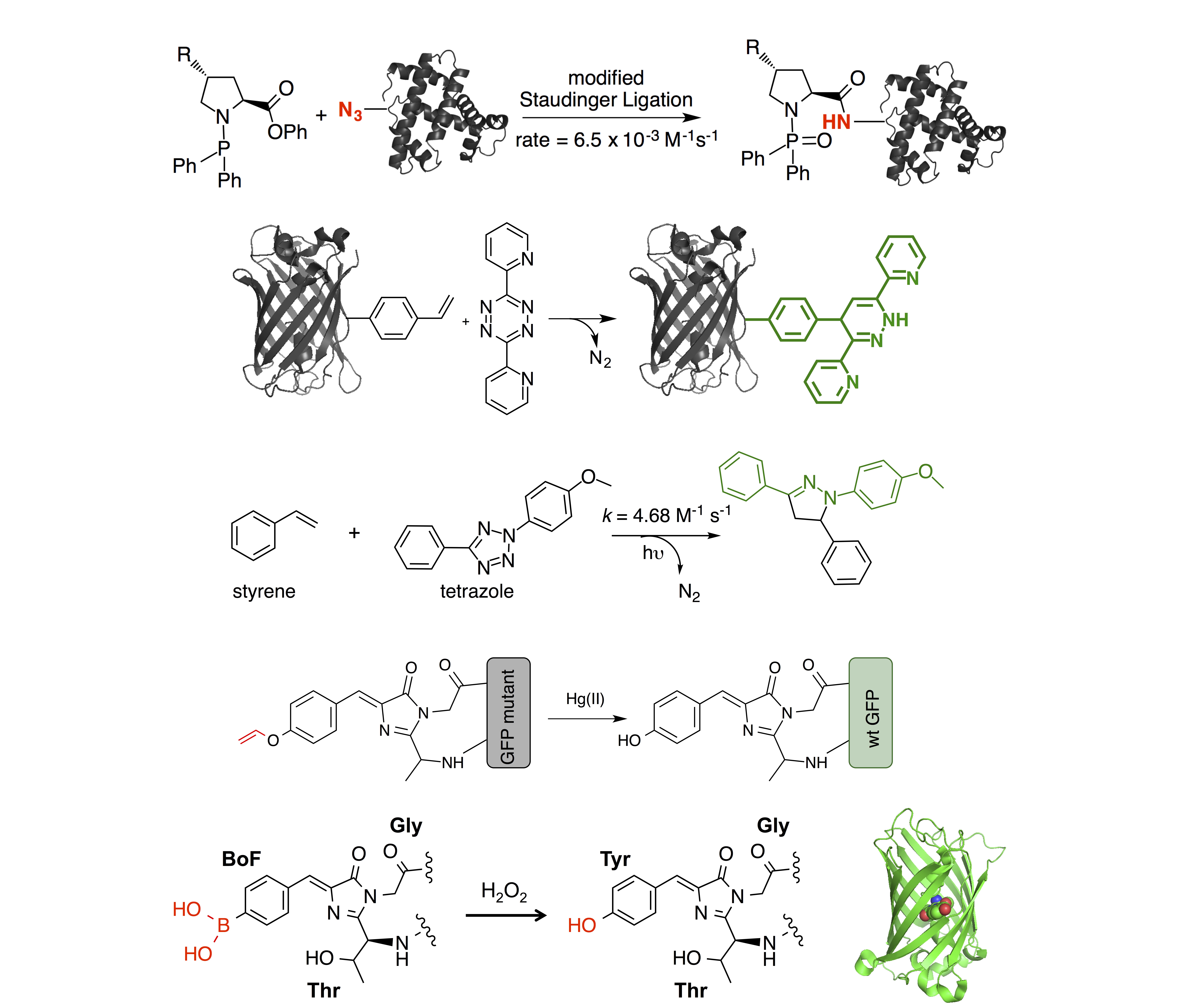
(a) Bioorthogonal reactions
The reassignment of a nonsense codon (e.g., amber UAG codon) into a sense codon has been widely explored for a programmed incorporation of unnatural amino acids into recombinant proteins. One focus of our research is to expand the scope of available aqueous phase chemical reactions so that a wider variety of biomolecules can be specifically labeled and/or probed in living systems. We are also interested in using caged amino acids to modulate protein function. While photo-decaging represents the most common strategy to release the caging group, my group seeks to develop additional decaging mechanisms that are orthogonal to each other and can be applied simultaneously to biological investigations. We are also interested in decaging mechanisms that can be triggered by endogenous stimuli (e.g., signaling molecules) of living organisms so that we can implement rationally designed strategies to either monitor or control a biological event of interest.
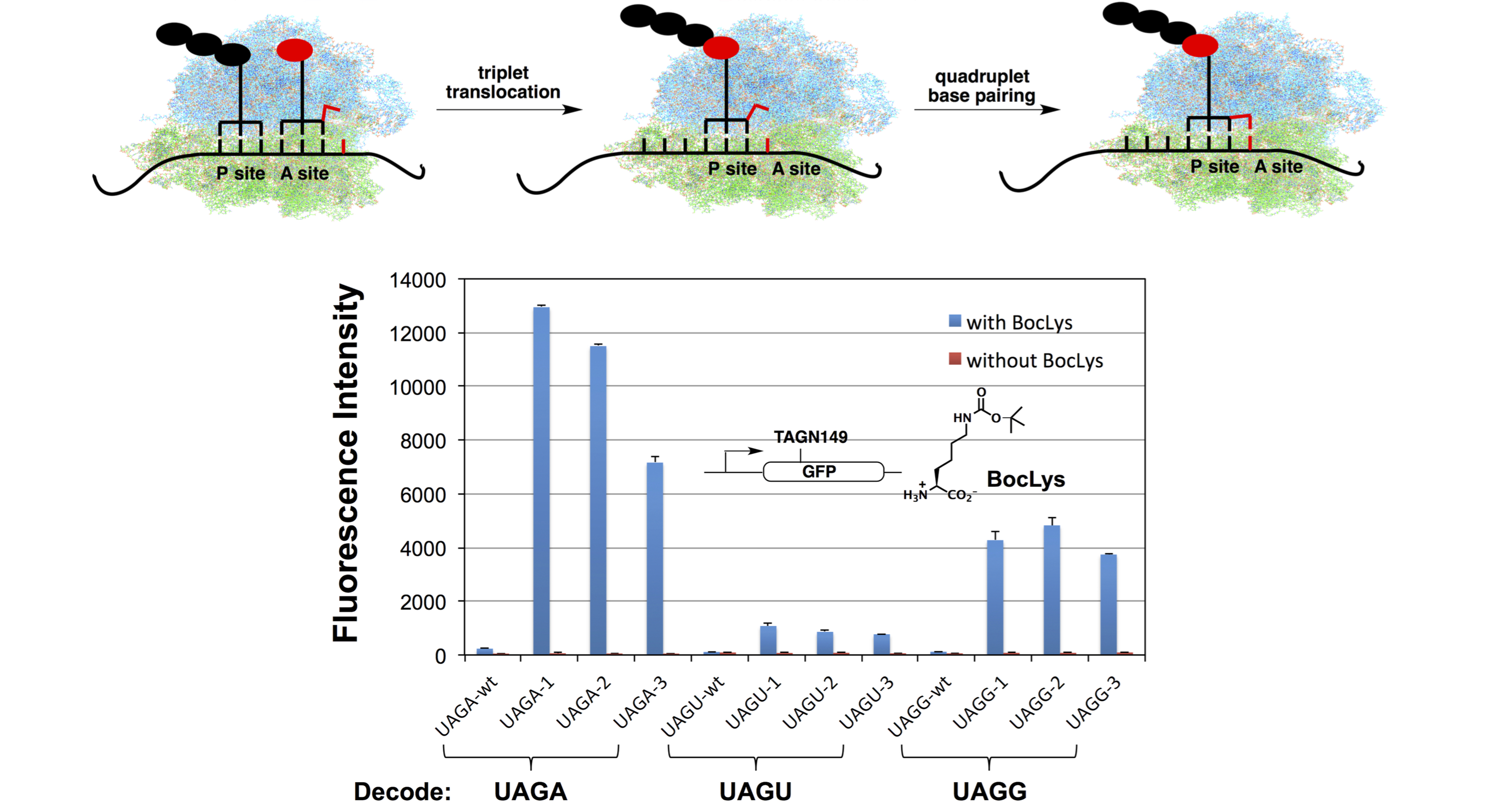
(b) Quadruplet codon decoding
While triplet codon is the predominant form of genetic code in nature, programmed frameshift, e.g., +1 frameshift (an apparent quadruplet codon decoding), in response to certain recoding signals embedded in the mRNA is a natural process. Recent developments in genetic code engineering demonstrated that quadruplet codon could be used to encode unnatural amino acids under experimental conditions in the absence of recoding signals. We recently reported a new approach, which is based on the engineering of tRNA anticodon stem-loop, to enhance the efficiency of quadruplet codon decoding. The introduction of quadruplet decoding mechanism provides additional blank codons (codons that do not encode natural proteinogenic amino acids), which potentially enable the genetically programmed functionalization of proteins with multiple unnatural amino acids in living cells. The potential of having diverse unnatural amino acids with novel physical and chemical properties in a single cell expands the scope of experimental designs for biophysical and biochemical studies. It also provides a platform to engineer cells with an increased biological diversity and enables novel design/synthesis of biological systems.
We are also investigating how mutations in tRNA mediate the switch from triplet to quadruplet reading frame within a universal translational machinery that prefers reading triplet codons. The inferences from our work will likely bring better understanding on how the ribosome maintains its reading frame, which remains one of the key questions to be understood in protein translation.
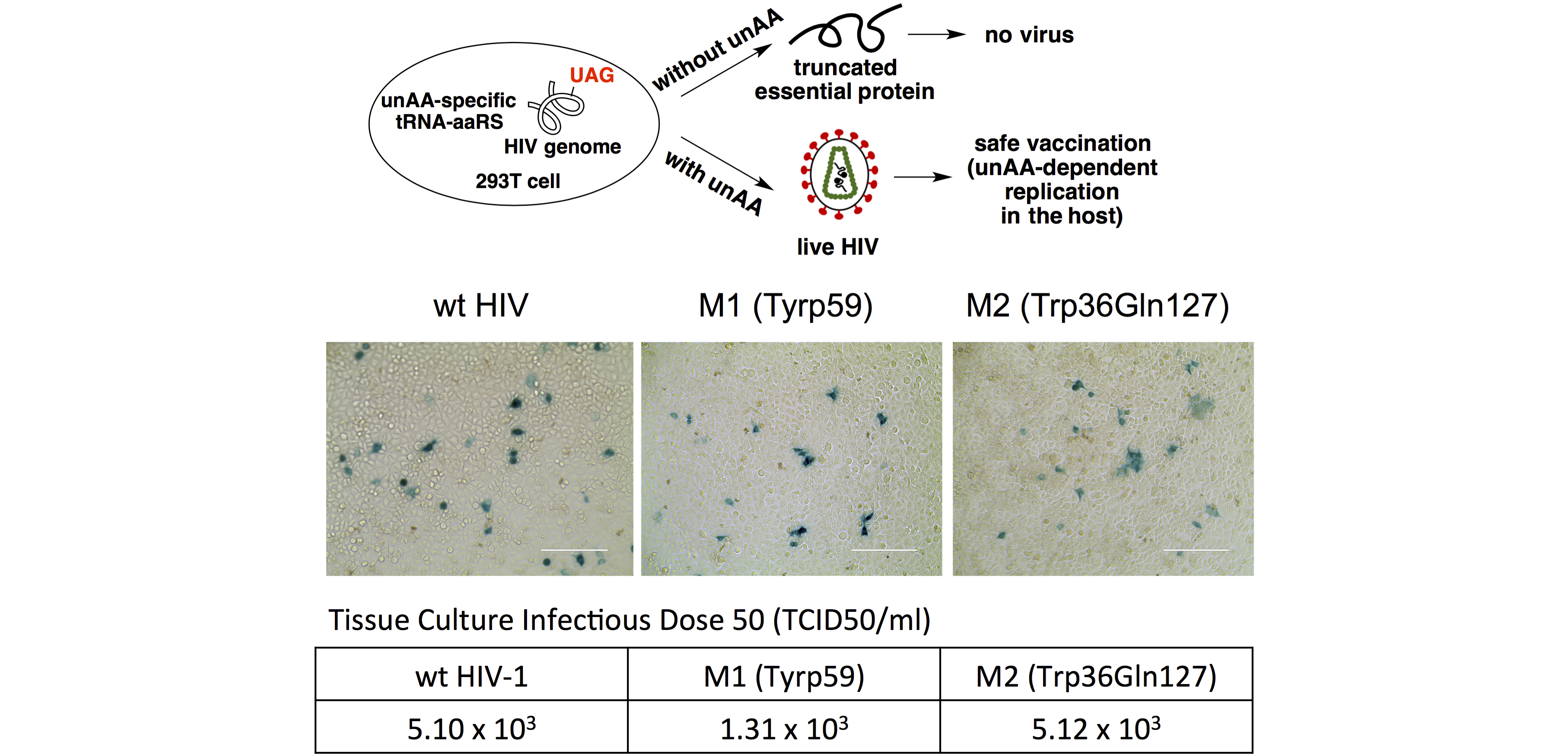
(c) HIV-1 Vaccine
A safe and effective vaccine against HIV is urgently needed to combat the worldwide AIDS pandemic, but it still remains elusive. Although live-attenuated virus vaccine showed excellent efficacy in eliciting host’s immune response, application of such strategy in HIV prevention is hindered by safety concerns due to uncontrolled replication of virus. My lab pioneered an innovative strategy in HIV vaccine development, which entails the manipulation of essential HIV protein biosynthesis through unnatural amino acid-mediated suppression of genome-encoded nonsense codon (PCT/US2015/015229, filed).
(d) Enzyme engineering with unnatural amino acids
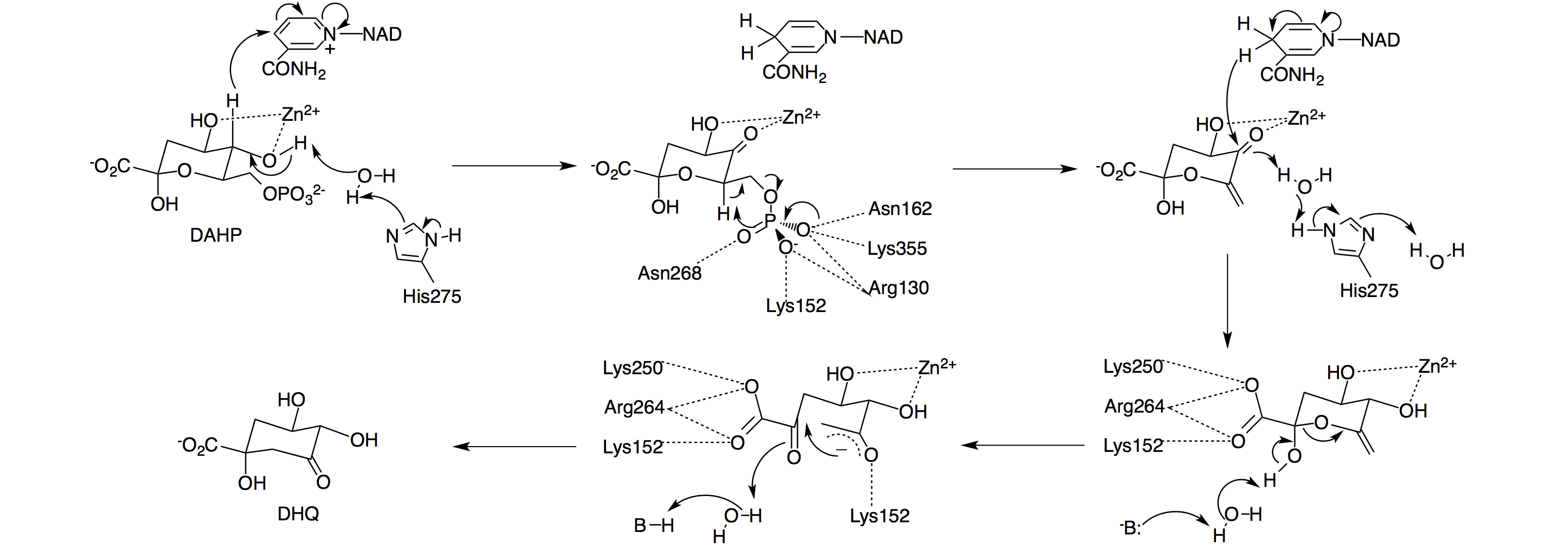
Directed evolution has become an indispensable tool to generate enzymes with improved properties, such as better catalytic activity and greater thermo-stability. Traditional directed evolution relies on the common twenty amino acids. The side chains of these twenty common amino acids comprise a relative limited number of functional groups. An intuitive question is if the addition of unnatural amino acids with novel physical, chemical, and biological properties to the genetic code can lead to the synthesis of proteins with enhanced properties. We are currently working on the evolution of an interesting enzyme, 3-dehydroquinate synthase, through unnatural amino acid mutagenesis. This enzyme is able to catalyze a complex multi-step reaction that includes oxidation, beta-elimination, reduction, ring opening, and intramolecular aldol condensation.
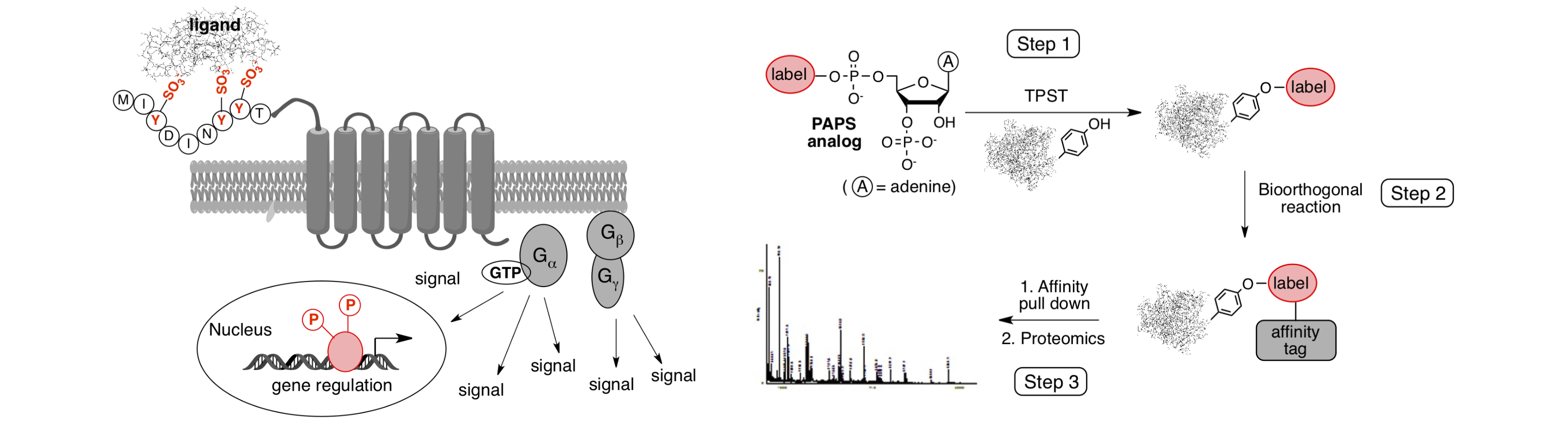
(II) Protein tyrosine O-sulfation
Protein tyrosine O-sulfation is considered the most common type of tyrosine modification in nature. It occurs exclusively on secreted and membrane-bound proteins that transit the trans-Golgi network. The introduction of negatively charged sulfate group plays crucial roles in extracellular biomolecular interactions that dictate various cellular processes including cell adhesion, leukocyte trafficking, hormone activities, and immune responses. Tyrosine-sulfated proteins also play important roles in the development of infectious diseases (e.g., AIDS and malaria), cancers, and a variety of immune-mediated diseases (e.g., allergy). Hence, protein tyrosine O-sulfation could emerge as an important therapeutic target for the treatment of human diseases.
We are developing chemical and biological tools to facilitate the study of protein tyrosine O-sulfation. We are especially interested in: (1) a discovery-based sulfoproteome study that lays the foundation to unveil the entire human sulfointeractome and to identify disease-related sulfoprotein-protein interactions; and (2) the role of sulfation of chemokine receptor in chemokine signaling, which is central to many chronic inflammatory conditions and involves in various steps of cancer carcinogenesis.
(III) Protein engineering and biocatalytic synthesis
(a) Molecular scaffolds
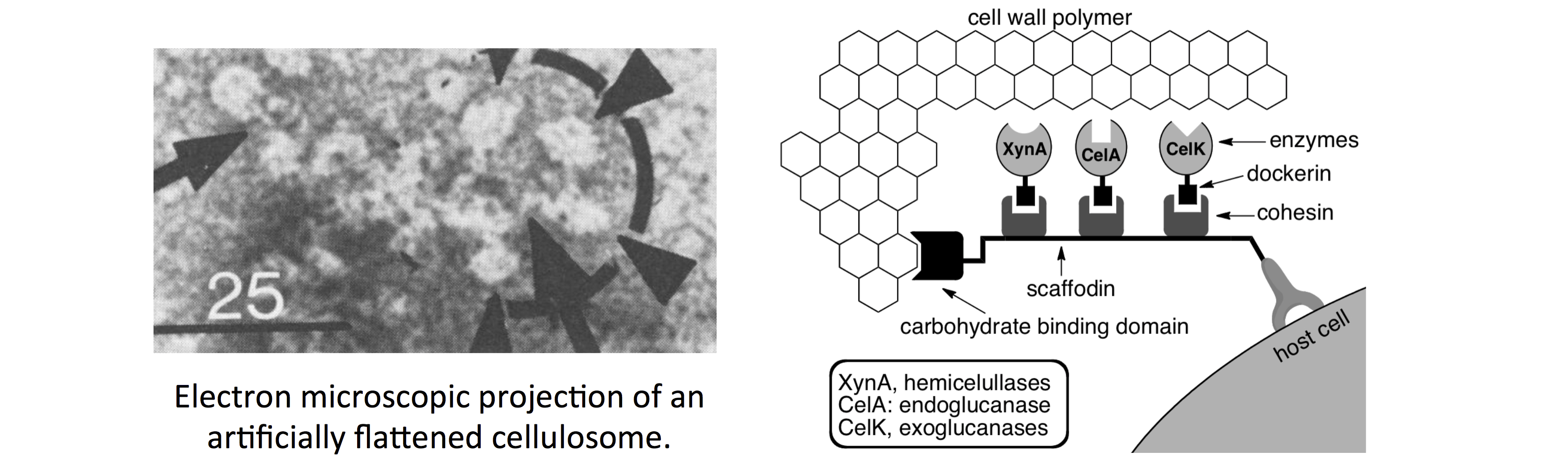
Energy is central to achieving goals for sustainable human society development. Due to heavy consumption and the nonrenewable nature, the current primary form of energy source, fossil fuels, will be depleted in a near future. As direct products of renewable raw materials, such as plant-based cellulosic material, biofuels represent a promising form of alternate sustainable energy. Large-scale production and commercialization of cellulosic biofuels requires a cost-effectively way to release soluble sugars from crystalline cellulosic material. Primary routes of plant cell wall deconstruction include biochemical conversion and thermochemical conversion. Our research focuses on improving the biochemical conversion of plant cellulosic material using the synthetic (designer) cellulosome, a protein complex in which key catalytic elements are attached onto a scaffold protein by virtue of the interaction between dockerin domains on the catalytic modules and cohesin domains on the scaffold. The co-localization of catalytic modules would potentiate synergistic activities among cellulosomal enzymes and result in more efficient degradation machinery than the free enzyme system. We have developed a general approach to obtain orthogonal cohesin-dockerin pairs with minimal crosstalking interaction, which is being applied to the regio- and stoichiometric controlled attachment of catalytic modules to achieve the assembly of synthetic cellulosomes with pre-defined molecular structure and catalytic property. The modular nature of synthetic cellulosome assembly also permits directed evolution in order to achieve higher cellulose degradation activities. This research is supported by NSF and has been integrated into the two graduate courses that I have been teaching. PowerPoint slides are available (click here).
(b) Biocatalytic synthesis
Chemical industry faces the sustainability challenges that arise from both the dwindling starting materials derived from fossil fuels and increasingly unaffordable environmental costs to the human society. As a solution to these problems, microbial syntheses are actively explored as the alternate platform of chemical production because of the smaller environmental footprint and microbial cells’ inherent ability to efficiently utilize renewable feedstocks as the starting material. In collaboration with Prof. Wei Niu at the Department of Chemical & Biomolecular Engineering, we seek to construct microbial catalysts in order to achieve efficient and scalable biosyntheses of value-added industrial or pharmaceutical chemicals.